What's new in CodonCode Aligner 7
CodonCode Aligner version 7.1 introduces support for two NGS tools from the BBTools package: NGS data trimming and adapter removal with Bbduk2, and NGS assembly with Tadpole.
CodonCode Aligner version 7.0 introduced functionality for RFLP analysis and improved NGS assembly.
Please note that CodonCode Aligner 7 requires a new license key. Version 7 is a free upgrade for all users with a current update and support agreement as of February 16, 2017. Other customers can purchase an upgrade to version 7.
RFLP Analysis
CodonCode Aligner 7.0 allows you to analyze your sequences for restriction fragment length polymorphisms.
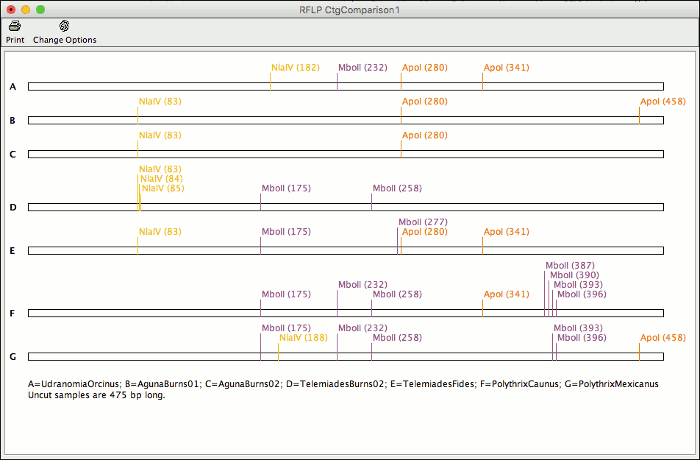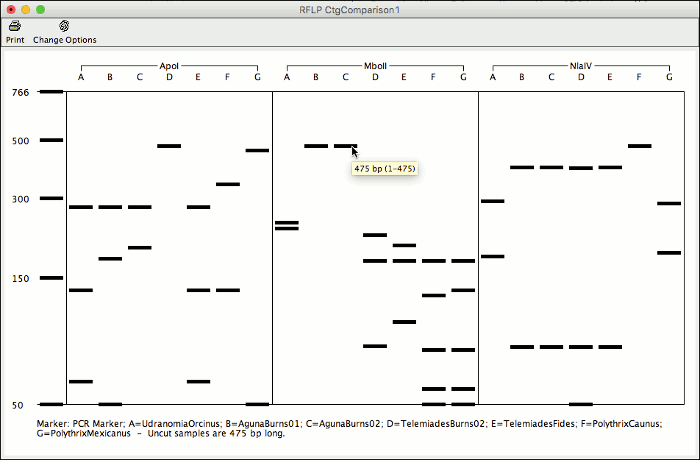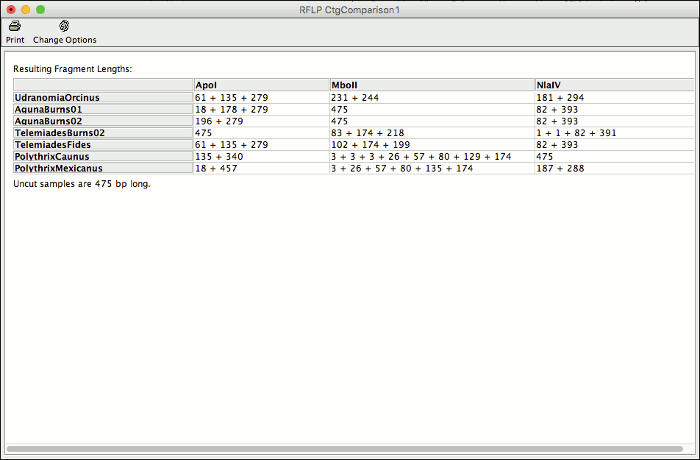
With this new feature you can compare cut sites for all samples in a contig and identify enzymes to use for your RFLP analysis. Generate a virtual gel of the expected results and print it to easily identify the lanes of your actual gel.
Aligner can automatically pick the enzyme that generates the most different cut result for your sequences. You can chane enzymes and display options for already generated results, or start a new analysis for the same contig to compare different results side by side.
|
Improved NGS Assembly
CodonCode Aligner 7.0 adds a new "built-in" assembly algorithm for NGS sequence data. Compared to NGS assembly with SparseAssembler (introduced in CodonCode Aligner version 6.0, and still supported), the new built-in NGS algorithm can give significantly larger contigs and N50 numbers. The following table illustrates this, using bacterial genome data from the GAGE and GAGE-B test sets:
Data set |
Read length |
SparseAssembler N50 |
CodonCode N50 |
SparseAssembler largest contig |
CodonCode largest contig |
Staph. aureus |
101 |
28,877 |
37,787 |
74,595 |
93,238 |
Vibrio cholerae |
250 |
2,726 |
80,570 |
19,705 |
207,445 |
Actual difference will vary depending on the data set. For longer reads like the 250 base Illumina data in the Vibrio dataset, contig sizes and N50 numbers can be more than 10-fold higher for the new built-in algorithm than with SparseAssembler.
Please note that NGS assembly in CodonCode Aligner is currently limited to small projects, for example bacterial genomes. Future improvements are planned. |
CodonCode Aligner 7.0 is compatible with Windows 10 and OS X 10.12 (Sierra).

