Sequence Editing with CodonCode Aligner
CodonCode Aigner provides many sequence editing tools, that allow manual as well as automated editing of your DNA sequences. You can edit your chromatograms in several different views, both before and after assembly into contigs. Sequence editing features include changing and deleting bases, reverse complementing, inserting gaps and bases, moving gaps and samples in contigs, undo and redo, deleting samples from contigs, splitting contigs, and changing bases to lower / upper case. For manual removal of longer sequence stretches at the start or end, CodonCode Aligner has "Delete from start" and "Delete to end" functions for both individual samples and contigs.
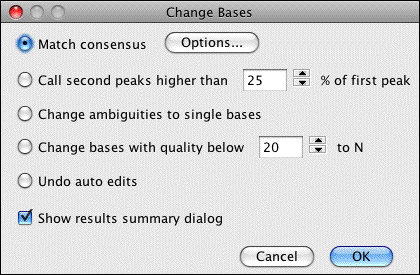
Several automatic sequence editing features are available through the "Change bases" submenu:
- Calling secondary peaks (using the corresponding IUPAC ambiguity code)
- Removing ambiguity codes
- Converting low-quality bases to Ns
- Automatically matching the consensus sequence
The screenshots below show an example of the "Call secondary peaks" feature:
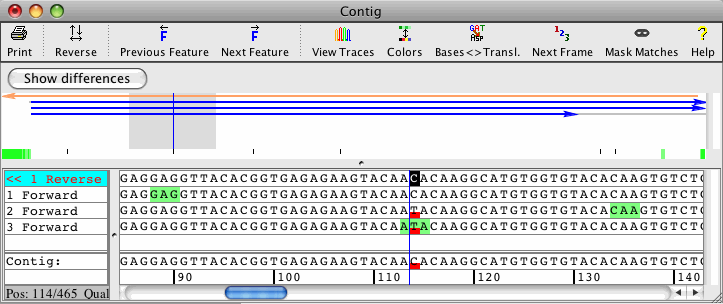
At the selected position in this assembly, two of our four sequences show the base C, the other two show the base T (in this example, the red box is used to highlight discrepancies). If you want see ambiguity codes at mixed peaks in this contig, you can use the "Change bases" feature and decide which secondary peaks should be called:
In the case shown above, the two Ts are changee to Ys:
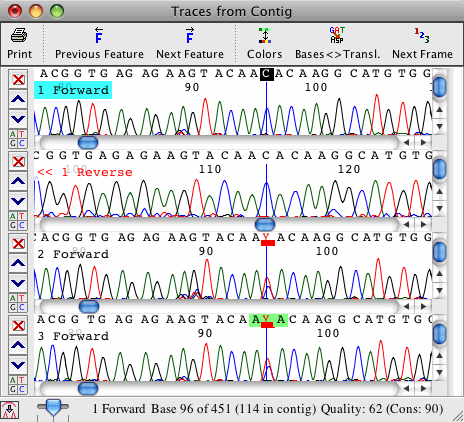
All automated edits can easily be undone at a later point in time, even after saving and closing a project, by using the "Undo auto edits" option.
