Screen Shots of CodonCode Aligner
This page shows screen shots of CodonCode Aligner, our tool for sequence assembly, alignment, editing, and mutation detection for Windows and Mac OS X. Click on any image for a larger view.
To see CodonCode Aligner in action, please visit the Tutorials page.
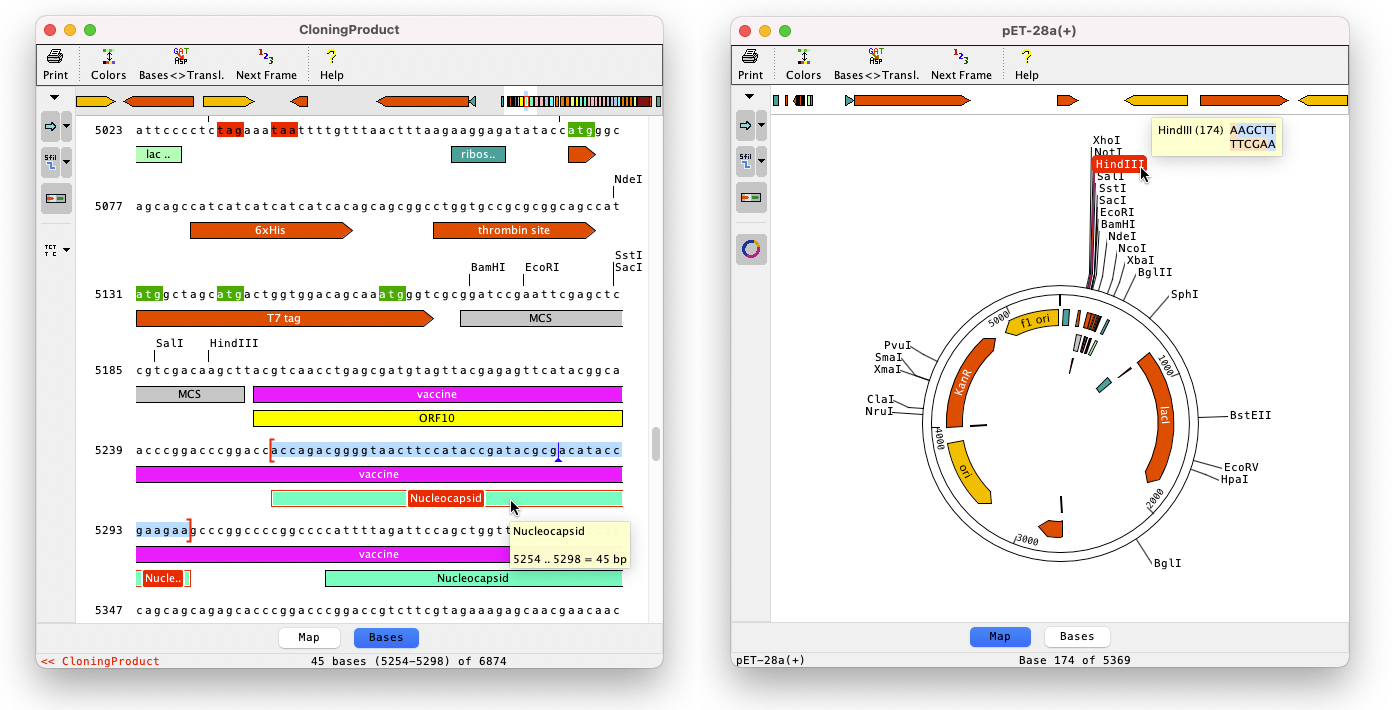
Base View: View Bases and Maps
The base view show bases or sequence maps. Bases and features can be edited. The user can customize which enzyme cut sites and features to show.
Clicking on features or enzymes allows to select specific regions for further processing, for example in our restriction cloning wizard.
Background colors can be adjusted to show things like translations or readign frames. Amino acid translations can be shown instead of bases.
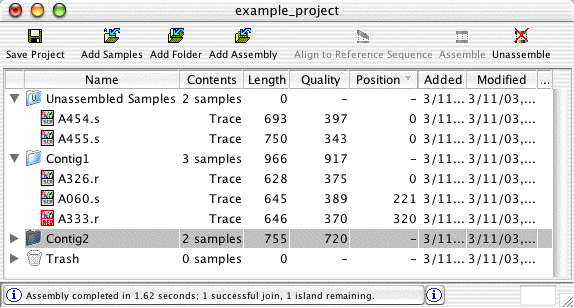
Project View: Organize Sequences and Contigs
The project view window shows the contents of projects - unassembled samples, assembled and aligned contigs, and the trash can. You can also create folders to organize your samples and contigs.
New sequences can be added to a project by dragging files or folders onto the project view. In the project view, samples and contigs can also be moved by drag and drop. For example, dragging unassembled samples onto a contig will add these samples to the contig (if they meet the minimum overlap criteria). Dragging a contig on the "Unassembled Samples" folder will unassemble the contig.
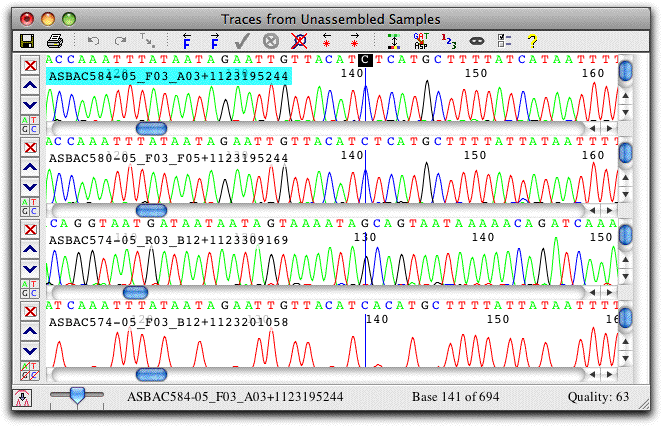
Trace View: View and Edit Traces
The trace view show sequence chromatograms and base calls. Traces can be scaled horizontally and vertically, and the traces for individual bases can be hidden, as shown in the lowest view.
The colors used can be adjusted to your preferences; traces can also be shown on a black background. Bases can be shown on a background that indicates sequence quality, a handy feature when examining discrepancies. Instead of the nucleotides, the amino acid translation can also be shown, and a translation-based background can be used.
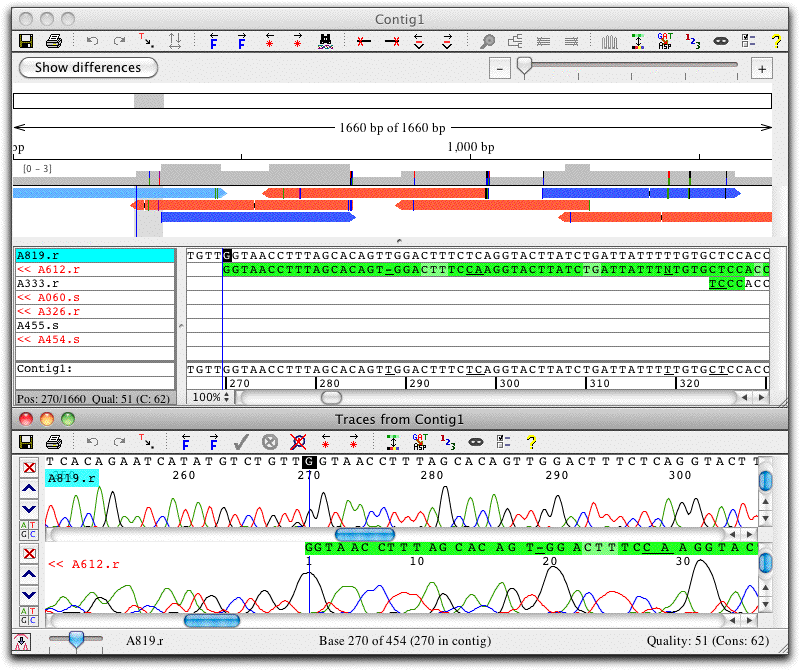
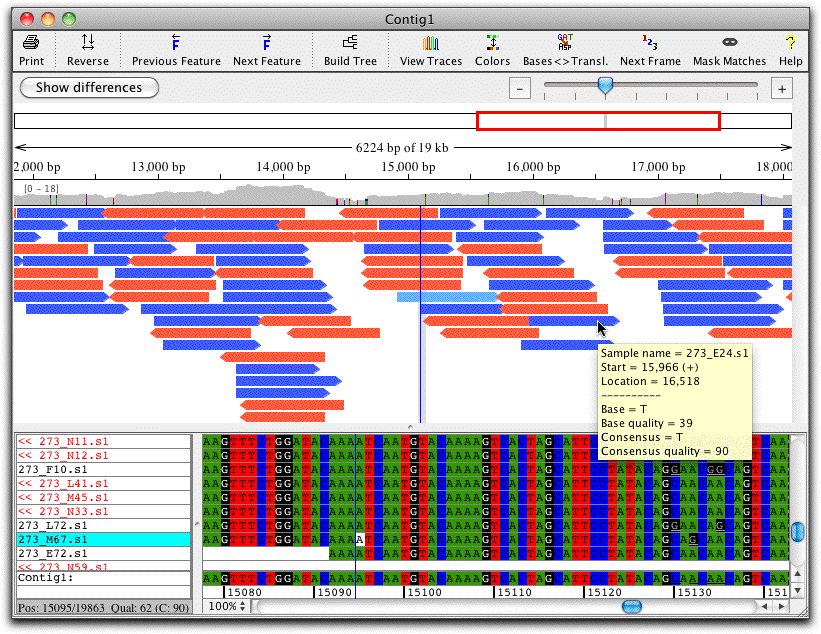
Contig View: Edit Assemblies and Alignments
The contig view window shows an overview of the contig at the top, and the aligned bases below. Double-clicking on bases in the contig view brings up the trace view, here shown below the contig view.
In this example, the green background indicates low quality. Note that the two reads in the section shown disagree at several places. However, the consensus sequence (shown at the bottom of the contig view) is correct even without any editing, since CodonCode Aligner uses the higher-quality sequence as the consensus sequence.
The toolbar at the top shows a number of buttons for quick navigation, e.g. to jump to the next discrepancy or gap (the buttons also have corresponding keyboard shortcuts and menu items). You can easily customize the toolbars in each window to show the icons that you use most.
Contig View: Overview of Your Contig
The contig view shows an overview of your contig at the top. The display of the sample arrows allows you to get a better idea of gaps and sample position in alignments. Mouse overs show significant information for each position, and when zooming in on the samples, discrepancies are shown in colors on the sample arrows.
The overview allows for zooming with a zoom scale and shows which range is displayed.
It also includes a coverage map above the sample arrows that shows colored discrepancies (with a settable threshold) and allows for fast navigation to any region of interest.
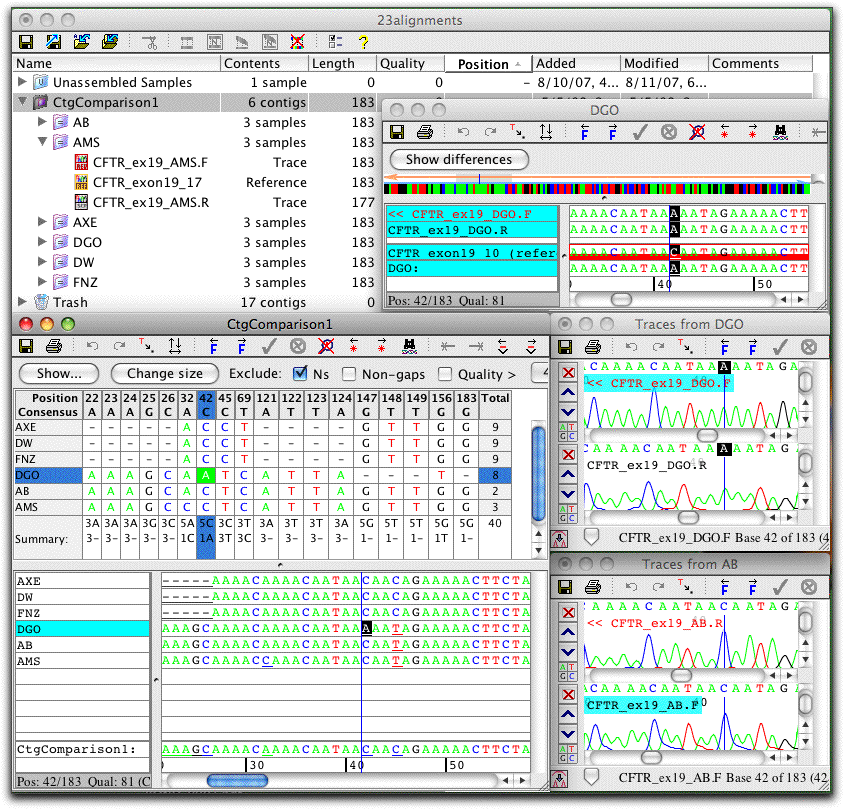
Align Contigs
A unique feature in CodonCode Aligner is the ability to build "contigs of contigs": to align contigs directly with the standard sequence alignment programs MUSCLE or ClustalW.
By aligning the contigs directly, rather than copies of the consensus sequences, the user keeps the option to go back with to the original sequence traces with a simple double-click.
An example of a contig alignment is shown on the screen shot to the left (click on the image for a larger view). Six contigs were aligned with MUSCLE to create a "contig of contigs", named "CtgComparison1". The contig view for this "parent" contig and the contig view for one "child" contig (named "DGO") is shown. Also shown are the traces for two contigs at this position (contigs DGO and AB).
Immediate access to the underlying sequence traces allows quick verification of differences. If necessary, edits can be made in any of the views; edits will be reflected right away in all the other views.
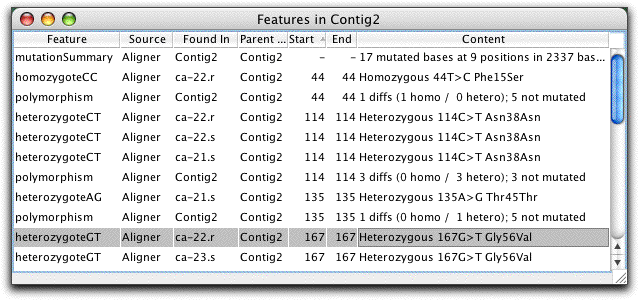
Feature View or: What Interests You?
The feature view window in CodonCode Aligner shows a list of "features" or "regions of interest". The user can define what exactly constitutes a feature - for example, regions of low coverage, or bases that were tagged as potential mutations when using Aligner's "Find Mutations" method. The example screen shot shows features for a SNP detection project. Note that the amino-acid level effect for each putative mutation is given.
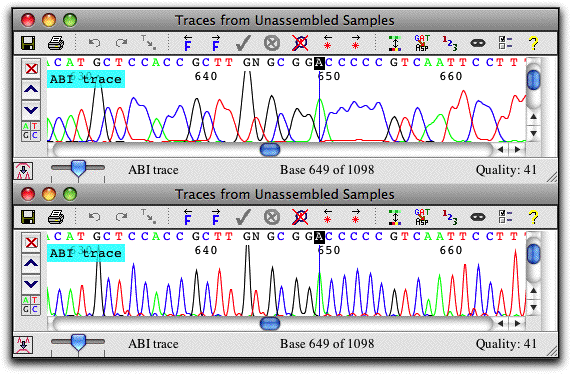
Trace Sharpening
Get a better look at your data with CodonCode Aligner's trace sharpening. If you need to work with "real world" sequences where the peaks start to overlap badly (top trace), just press a button, and CodonCode Aligner will generate a sharpened version of the trace, as shown at the bottom.