Barcoding Insect Species from Pakistan - A CodonCode Aligner User Showcase

At the National Institute for Biotechnology and Genetic Engineering in Pakistan, CodonCode Aligner is used to barcode insect species from Pakistan. This is a collaborative project between Dr. Muhammad Ashfaq's lab in Pakistan and the Biodiversity Institute of Ontario (BIO) Canada where CodonCode Aligner is the favorite tool to edit, assemble, align and analyze barcoding sequence data.
"A significant number of sequence files are generated when barcoding a large number of insect species. Having access to this software will facilitate my barcoding work and I will be able to analyze and upload the barcode data at the Barcode of Life Data base efficiently. I saw people working at BIO with CodonCode Aligner and found this software very good. DNA analysis done at my lab will become compatible with that of BIO." says Dr. Ashfaq about being able to use CodonCode Aligner for his research analysis.
CodonCode Aligner is the favorite software used to analyze data in the iBOL (International Barcode of Life) project. Lead DNA Scientist Natalia Ivanova, Ph.D. from the Canadian Centre for DNA Barcoding at the Biodiversity Institute of Ontario, University of Guelph says: "The iBOL project is growing rapidly and the DNA barcoding facilities are being established all around the globe. One of the key elements is the CodonCode software. In my opinion, this is one of the best solutions for assembly of DNA barcodes. We often recommend this software to our colleagues and visiting scientists."
Dr. Ashfaq and his research associates are now using CodonCode Aligner for four different projects, all of which involve gene isolation, sequencing and sequence analysis:
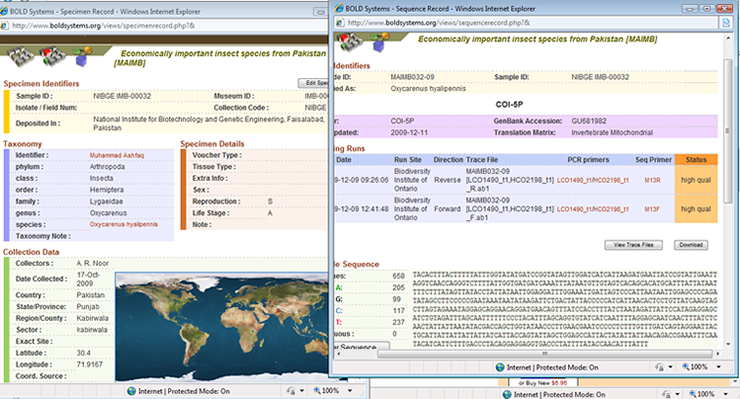
- Sequencing DNA barcodes of economically important insect species of Pakistan.
- Molecular diagnostics of subterranean termites of Pakistan.
- Molecular basis of insecticidal resistance and estimation of insecticide resistance allele frequency in cotton bollworm populations in Pakistan.
- Gene silencing and functional genome studies of cotton mealybug by RNA interference: implications in insect control.
Sequencing DNA Barcodes of Economically Important Insect Species of Pakistan
by Muhammad Ashfaq

Correct identification of living organisms has always been a challenge. Insects are not an exception to this difficulty. There are millions of insect species, some are very conspicuous and larger in size and some are concealed and very minute. Developments in the science of molecular biology have provided a new direction and insight in the identification of organisms and to separate them from their closest relatives, which was otherwise very difficult. Some new species have been discovered based on the differences in the genome sequences. Insects are the major player in incurring economic losses to crops and fruit trees in Pakistan. Some insect species have been fully explored and identified while the identification of some species is ambiguous. Identification of insect species through DNA sequencing and developing a sequence database and reference library can be very productive for the agricultural scientists in general and taxonomists and entomologists in particular. DNA barcoding is an idea and approach which is based upon the utilization of nucleotide sequence differences among individuals to identify them to species or race level. The project is part of a broader approach taken through the International Barcode of Life (iBOL) project to barcode millions of species from across the world. National Institute for Biotechnology and Genetic Engineering (NIBGE) is contributing in the international effort directly by barcoding local insect species from Pakistan. On one hand the project will contribute internationally and on the other hand it will benefit the country itself by DNA barcoding the local insect species. Insects of economic importance (both pests and beneficial) are being collected from different geographical localities of Pakistan. We are targeting both the nuclear (18S, 28S, EF-1a and ITS1 and ITS2) and mitochondrial (COI and COII) genes for sequencing. Work is being done in collaboration with the Biodiversity Institute of Ontario (BIO), Canada. CodonCode Aligner is useful for assembly and sequence alignments, sequence editing, end clipping, translations and so many other features required for sequence analysis.
Molecular Diagnostics of Subterranean Termites of Pakistan
by Muhammad Ashfaq
Termites are a big problem in both the agricultural and urban areas of Pakistan as they cause significant damage to crops and buildings. More than fifty species of termites have been reported from Pakistan and there may be many more. We are collecting termite samples from various geographical localities of Pakistan to identify the species through DNA sequence data. We use mitochondrial (COI and COII) genes for species identifications and Internal Transcribed Spacer (ITS) regions (ITS1 & ITS2) to study genetic diversity among and within termite species. This work is generating a large number of nucleotide sequences which needs to be analyzed by reliable software.
Molecular Basis of Insecticidal Resistance and Estimation of Insecticide Resistance Allele Frequency in Cotton Bollworm Populations in Pakistan
by Muhammad Ashfaq

(Image by Gyorgy Csoka, Hungary Forest Research Institute)
A number of factors are responsible for development of resistance in insect populations in the agricultural system. In order to suppress or delay the development of resistance and establish a reliable resistance monitoring system, it is important to understand the mechanism of insecticide resistance. Target-site resistance (knock-down) to pyrethroids and metabolic resistance to all insecticides are major sources of insecticidal resistance. Target-site resistance to pyrethroids and to cyclodienes is controlled by changes in regulatory genes determining the number of receptor protein molecules synthesized. The major gene involved in knock-down resistance against pyrethroids in insects is sodium channel gene. Metabolic resistance is achieved mostly by detoxification of insecticides through a family of genes called cytochrome P450s or some uncharacterized genes. In this project insecticide-resistant and – susceptible strains of cotton bollworm have been established in the controlled conditions and already reported and novel genes involved in both metabolic and target-site resistance are being cloned and sequenced. Expression of the sequenced genes is evaluated in response to different groups of insecticides to determine the role of a particular gene against a particular insecticide. Conserved point mutations are located in resistant insects and protocols are developed for PCR amplification of specific allele (PASA) for assessment of resistant genotypes present in the field. Bollworms collected from different localities from cotton growing areas of Pakistan are used for amplification of genes involved in resistance. Resistant genotypes and resistance allele frequencies are determined by PASA and compared between heavily sprayed, moderately sprayed and minimal sprayed localities. Findings will be useful in devising new insect management strategies on agricultural crops in general and cotton crop in particular. In this project CodonCode Aligner can primarily be used for sequence assembly and alignments, locating point mutations, end clipping and trimming vector sequences.
Gene Silencing and Functional Genome Studies of Cotton Mealybug by RNA Interference: Implications in Insect Control
by Muhammad Ashfaq

(Photo by M. Ashfaq, NIBGE, Faisalabad, Pakistan)
Research on functional genomics of insects can produce tangible results by providing directions for modern insect pest control such as through gene maneuvering and development of transgenic plants. Numerous identified genes in insects are functionally uncharacterized, which is a challenge to our understanding of insect behavior and development. Gene expression and protein synthesis in living organisms is regulated and maintained by various means. One known mechanism for gene regulation is post transcriptional gene silencing through degradation of mRNA by double stranded RNA (dsRNA) and is called RNA interference (RNAi). RNAi has been tested so far in a number of organisms and can be used as a tool to investigate the in vivo localization and precise role of genes in insects. Major agricultural crops in Pakistan in general and cotton crop in particular are severely damaged by a number of insect species. Recently cotton mealybug, Phenacoccus solenopsis a sap-sucking insect has appeared as a major threat to cotton crop.
The studies in the project have focused on the functional genomics of mealybug. Genes which play a key role in insect development, survival and insecticidal resistance are identified and isolated, and their specific functions are determined. The research will yield knowledge much needed to understand the insects at gene level. The information will pin point the potential target genes which could be incorporated in future insect control strategies such as insect control through RNA interference or through transgenic plants. Thus, in addition to gene function studies, RNA interference may also prove a tool for insect control through gene suppression. In this project CodonCode is being used for sequence assembly, end clipping, trimming vector sequences, translations and several other functions.
