What's new in CodonCode Aligner 10.0
CodonCode Aligner version 10.0 introduces restriction cloning, sequence maps, support for separating alleles, and more.
Please note that CodonCode Aligner 10 requires a new license key. Version 10 is a free upgrade for all users with a current update and support agreement as of December 17, 2021. Other customers can purchase an upgrade to version 10.
Version 10.0.2 fixes several bugs, and offers improved accuracy when separating alleles.
Restriction Cloning
In CodonCode Aligner 10 you can do virtual restriction cloning.
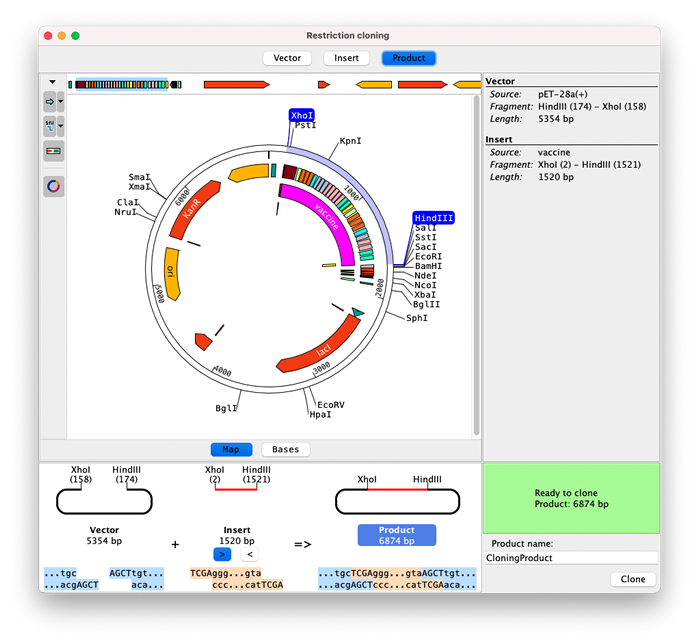
The vector and insert sequences can be displayed as linear or circular maps, or on a base level. Enzymes and sections to clone are easily selected by clicking on enzymes or features.
The bottom section shows how vector and insert fit together and if a product can be formed based on the cut sites.
The toolbar buttons on the left of the view contain options to change which map, features and enzymes are shown, for example showing only enzymes that cut once.
|
Sequence Maps
Tying in with the cloning feature, Aligner 10 adds the possibility to show sequence maps. Circular and linear maps can be shown, and they display customizable features and enzyme cut sites.
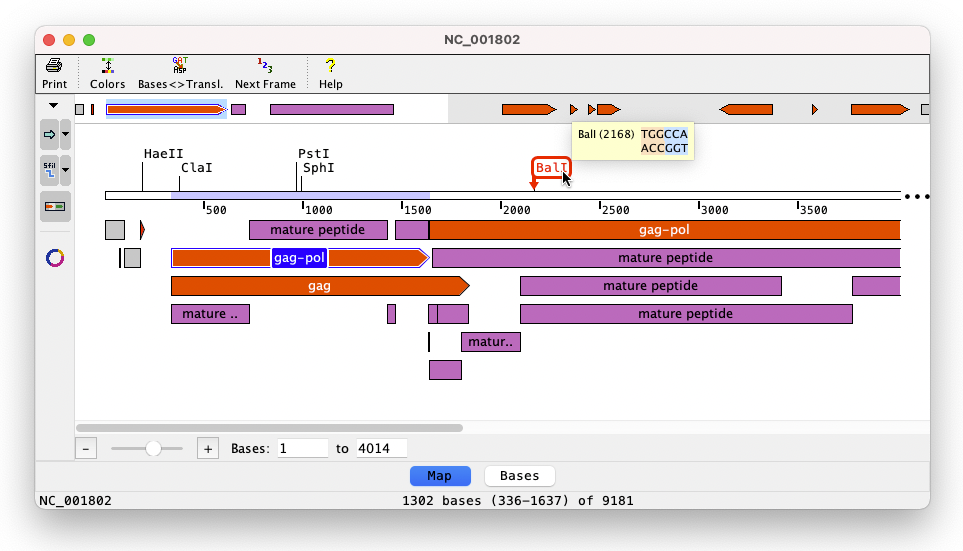
The mini-map at the top gives an overview. Selecting a region in the sequence is easy by clicking on a feature or shift-clicking on two enzymes. Mouse overs show detailed information for cut sites and features. And a zoom option at the bottom allows zooming in on specific areas. You can quickly switch between map and bases after selecting or naviagting to a specific region, and the buttons in the toolbar on the left customize map options like which features and enzymes are shown.
|
Separate Alleles
The "Separate Alleles" function in CodonCode Aligner is intended identify linkages between alleles in small NGS sequencing projects from diploid organisms. Here is an example that shows linked alleles in paired end sequences:
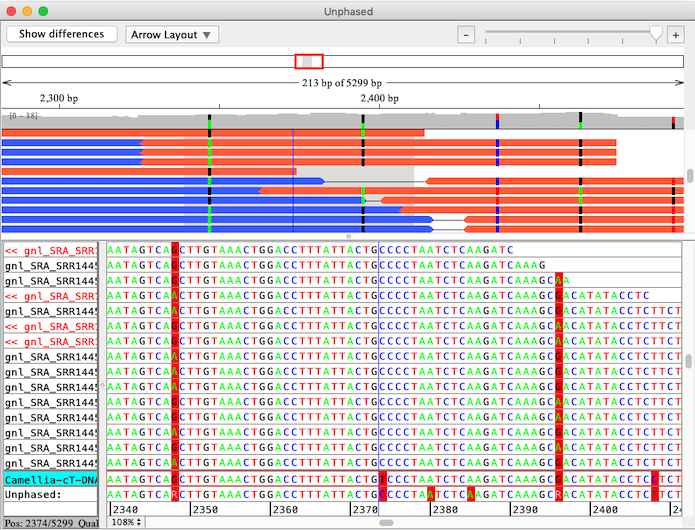
CodonCode Aligner uses a multi-step process to align sequences to a reference sequence, identify linked alleles, and separate sequence pairs into linked groups. The final results of the analysis are individual contigs for each group of linked alleles, which are then aligned to the original reference sequence using CodonCode Aligner's "Compare contigs" function. The screen shot below shows the result of such an analysis, with the "contig of contigs" allele alignment on top, and the allele-specific contigs below:
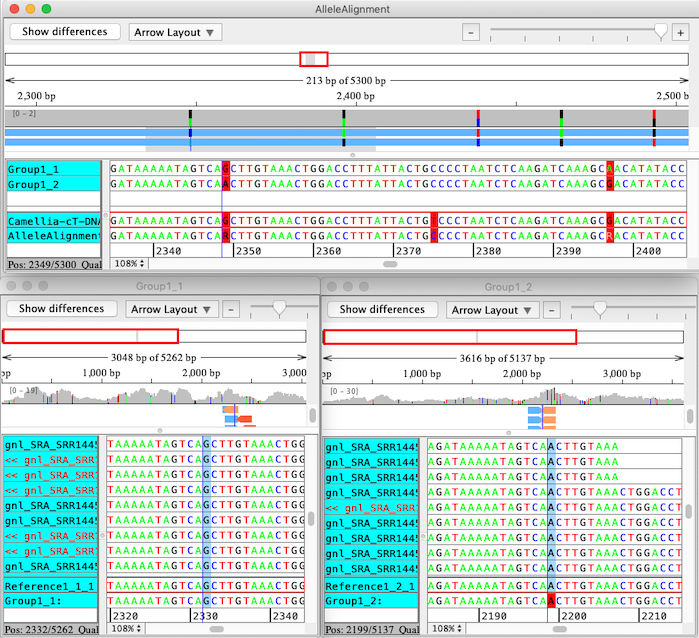
|
More...
- Show single letter amino acid translation on base level
- Highlight start and stop codons in sequences
- Show bases in customizable groups or without grouping
|
CodonCode Aligner 10.0 is compatible with Windows 11 and macOS Monterey.
Interested? Watch the movie showing the new features of CodonCode Aligner 10.0.1.




