Mutation Detection
CodonCode Aligner offers several features and methods for the detection and analysis of heterozygous point mutations:
- Calling secondary peaks in sequence traces
- Highly sensitive SNP detection by analyzing aligned traces in contigs
- Methods for analyzing sequence traces with heterozygous insertions or deletions (indels)
For heterozygous point mutations, CodonCode Aligner analyzes traces for the presence of both a secondary peak and the reduction in intensity of the primary peak. This approach is (a) more sensitive and (b) less likely to yield false positive results than simpler approaches that analyze only secondary peaks.
CodonCode Aligner can use open reading frame annotation from reference sequences to give amino-acid level annotation of the effect of point mutations:
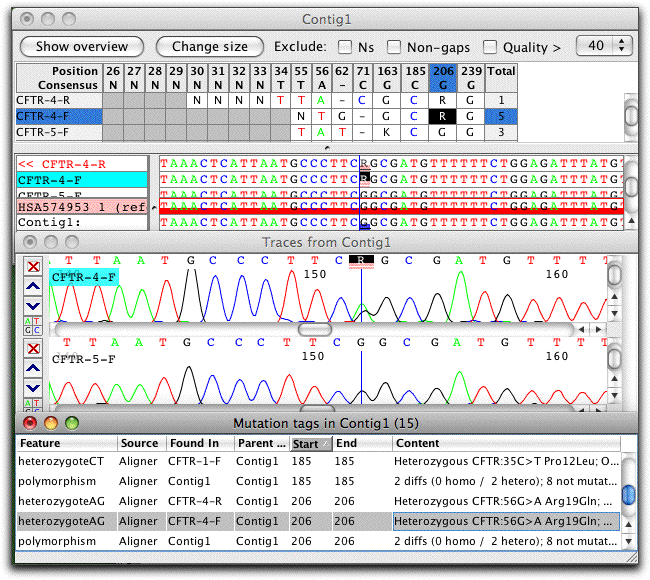
CodonCode Aligner also provides several methods for the detection and analysis of heterozygous indels. Here is an example of sample with a heterozygous two-base deletion that was separated into two pseudo-alleles with CodonCode Aligner's "Split Heterozygous Indels" option:
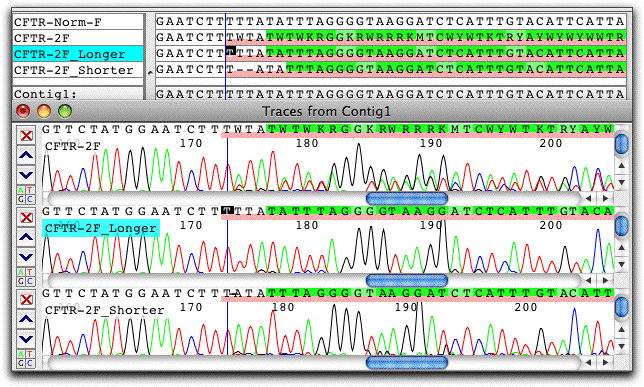
The "shorter" pseudo-allele trace created by CodonCode Aligner clearly shows the deletion of two Ts.In addition to the splitting method shown above which does not require any additional information beside the sequence chromatogram, CodonCode Aligner offers two additional methods to analyze heterozygous indels within the contect of contigs.
