Sequence Translation with CodonCode Aligner
CodonCode Aigner provides several options for viewing amino acid translations of your sequences. You can show the protein translation for every sequence in your project, as well as for your consensus sequences. Here is an example of the contig view where the sequence translations and the consensus translation for the forward three reading frames are shown:
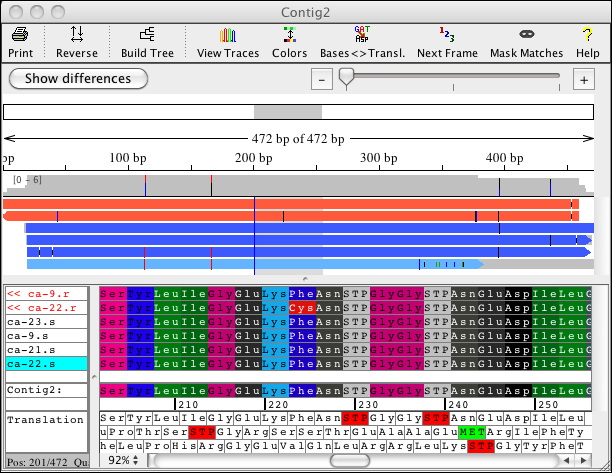
You can switch between showing the bases and the amino acids when you look at your sequences. For the sequence amino acid translation, you can choose one of the three possible frames. The protein translation for your samples can also be shown by using background colors. This allows you to look at your bases while seeing the amino acid translation as colored background, or looking at the amino acids and having the background colored by base:
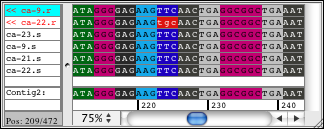
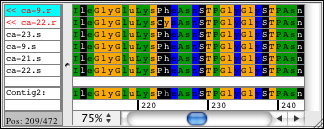
CodonCode Aligner can also show a protein translation of your consensus sequence for annotated coding regions. Typically, this display is used together with reference sequences imported from Genbank or EMBL-formated files; CodonCode Aligner uses the coding sequence annotation from these files if it is present.
You can choose between no translation, the translation of one reading frame, all three forward frames, all six forward and reverse reading frames, and annotated coding regions. Start and stop codons are highlighted by colors and amino acids names are represented by single- or three-letter codes.
The organism for which the sequence translation is being performed can be selected in the CodonCode Aligner preferences.
